Conventional diagnostic methods for detecting Salmonella in live poultry production typically rely on analyzing isolated bacterial colonies from enriched samples.
Conventional diagnostic methods for detecting Salmonella in live poultry production typically rely on analyzing isolated bacterial colonies from enriched samples.
Recent evidence, however, suggests these methods can overlook the presence of certain strains and serotypes, underestimating the true prevalence and risk of Salmonella in a sample, says Margaret Kiss, PhD, director of R&D at Ancera.
To effectively reduce Salmonella risk in a poultry-production system, she says, companies need accurate intelligence on:
- Detection: Is Salmonella present?
- Quantity: How much Salmonella is present?
- Strains and serovars: What Salmonella strains and serovars are present?
- Susceptibility: What kills or inhibits it?
“Biases such as bacterial-growth kinetics, variations in selective enrichment media and response to antimicrobial interventions widely differ across Salmonella serotypes and strains and can impact our awareness of food-safety threats,” Kiss explains.
Differences in growth kinetics
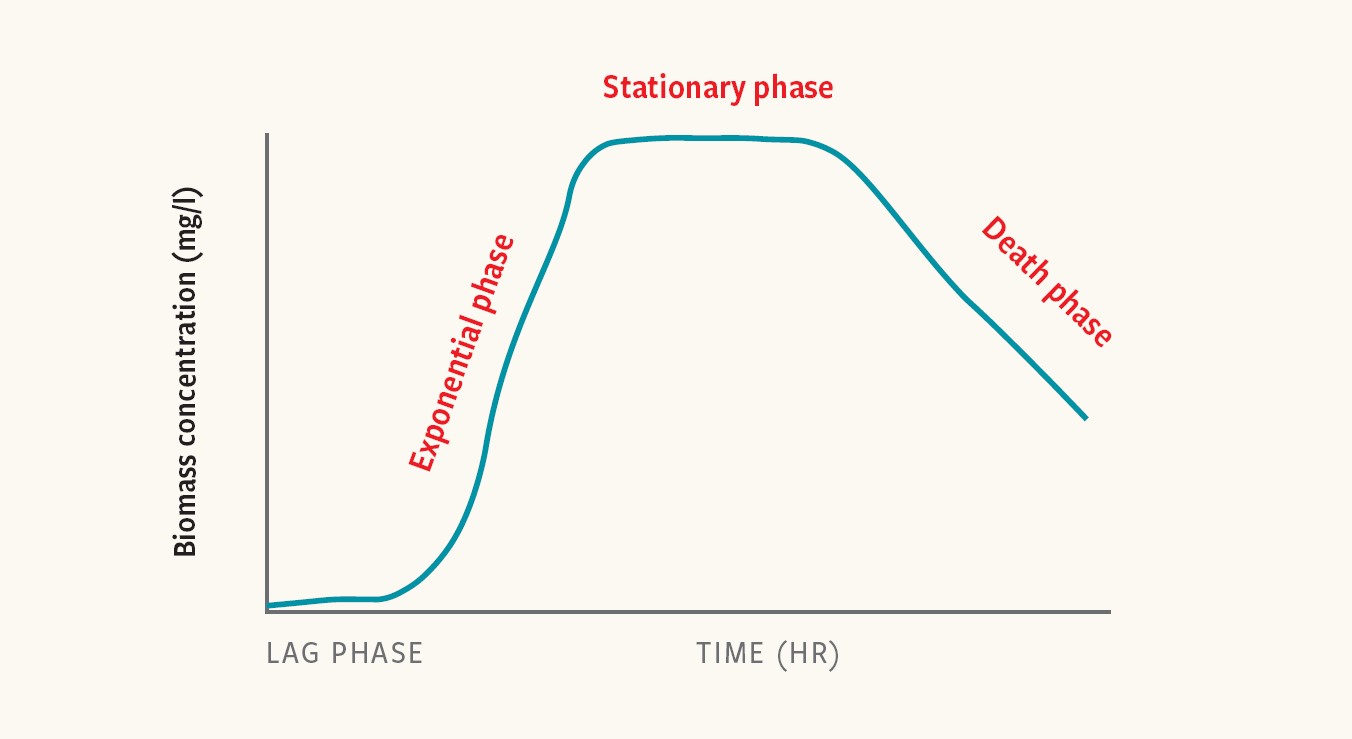
Bacterial-growth kinetics are often described as having four phases: the lag phase, exponential phase, stationary phase and death phase.
Each is characterized by distinct growth rates and physiological states. Different Salmonella strains (genetic variants) and serovars (distinct variations within a species) exhibit different growth kinetics, which can impact the detectability of Salmonella in enriched samples.
Figure 1. Comparison of unstructured kinetic bacterial-growth models
“Unique characteristics in the length of the phases will impact the number of cells present after a given enrichment time,” Kiss adds.
Serotype and strain-specific differences in lag time
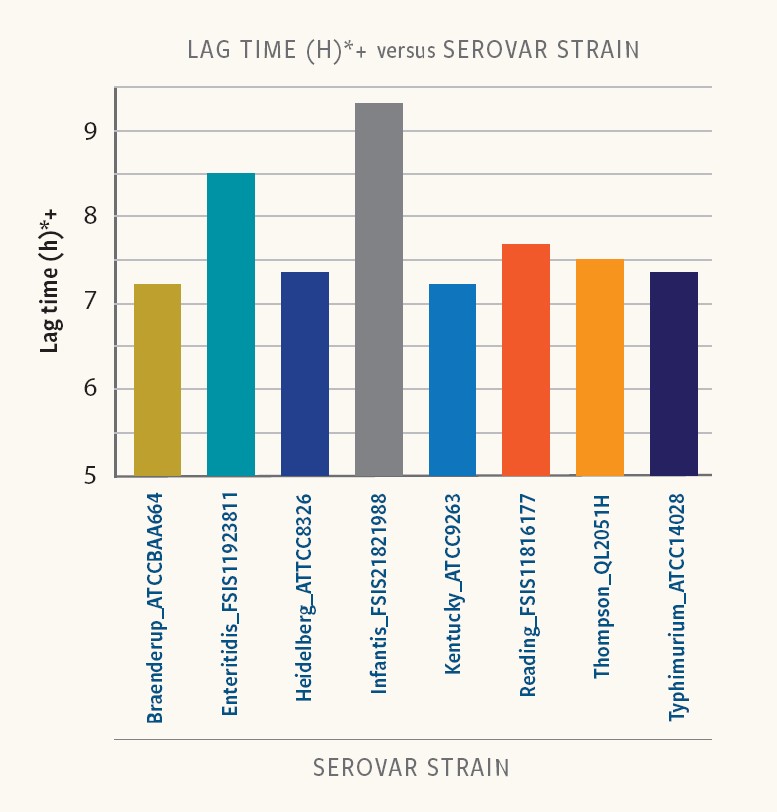
Different Salmonella serovars and strains exhibit considerable variability in duration of the lag phase.
Figure 2. Lag time in strains of different serovars
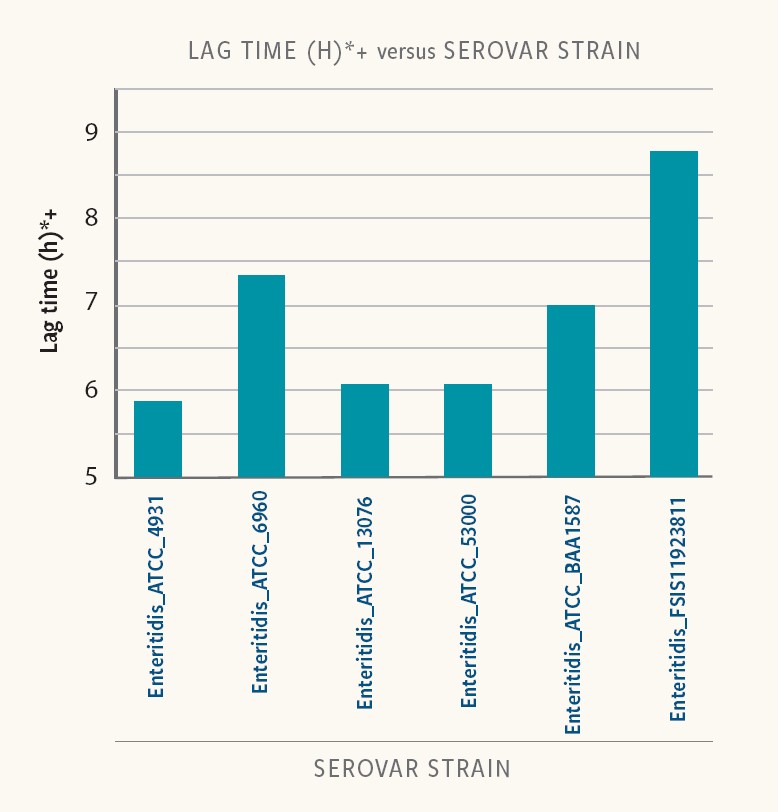
Figure 3. Lag time in different strains of the same serovar
Ancera’s internal research shows that lag time varied from 6 to 9 hours at low-starting inoculum (10-100 cfu/ml) in buffered peptone water (BPW). Even strains of the same serotype exhibited variability in lag times.
“Food-safety experts must incorporate differences in strain-specific lag time to properly interpret the results of Salmonella isolation and quantification,” Kiss says.
Doubling-time variability
Doubling time — the amount of time for a bacterial population to double in size — also has significant variability among different Salmonella strains and serovars.
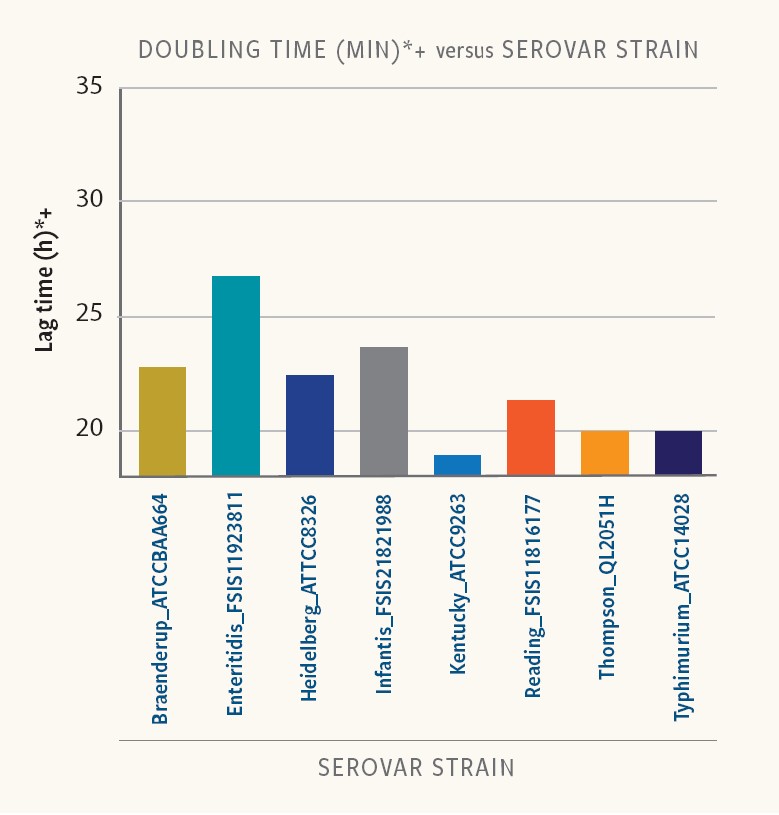
Figure 4. Doubling time in strains of different serovars
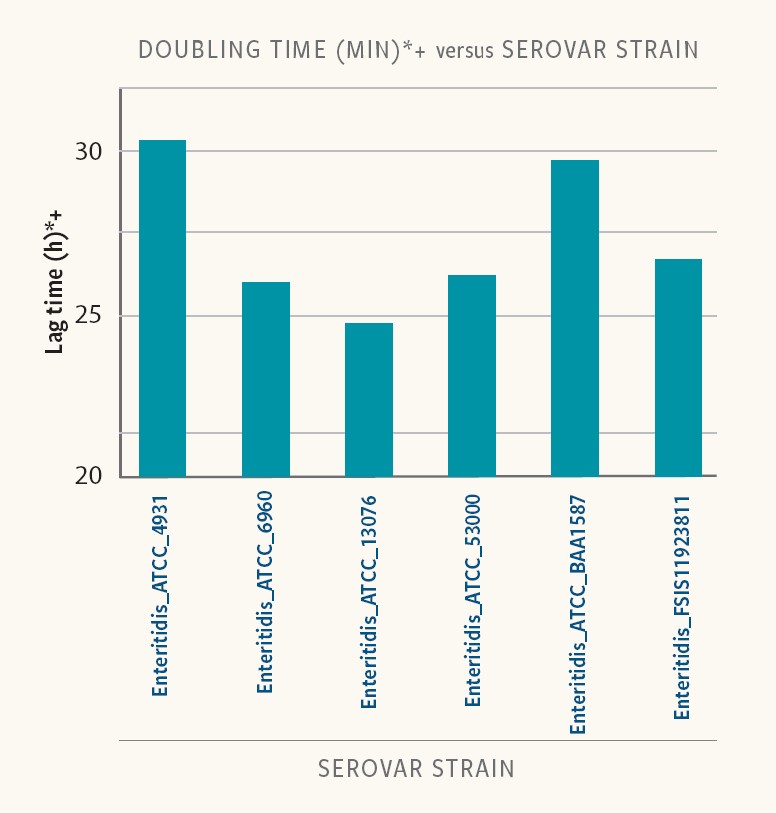
Figure 5. Doubling time in different strains of the same serovar
In Ancera’s research, doubling time varied between 19 and 30 minutes for the different strains and serovars examined in BPW.
“And in addition to differences in lag times, different strains of the same serotype had different doubling times,” Kiss reports. “For this set of strains and growth conditions, a quantitative Salmonella measurement could be impacted in a mixed serovar population.”
For example, after 9 hours the Salmonella Kentucky population would have gone through about six doublings while the Salmonella Infantis population would, on average, still be in lag phase. Accordingly, S. Kentucky would be over-represented in the sample relative to S. Infantis after enrichment.
Selective media in Salmonella surveillance
Selective media facilitates the isolation and identification of target organisms from samples. To help with detection, these media typically contain ingredients that inhibit the growth of non-target bacteria while allowing the growth of Salmonella.
“However, the culture media can influence the Salmonella-growth kinetics depending on the composition of the media and the characteristics of the Salmonella strains present in the sample,” Kiss cautions.
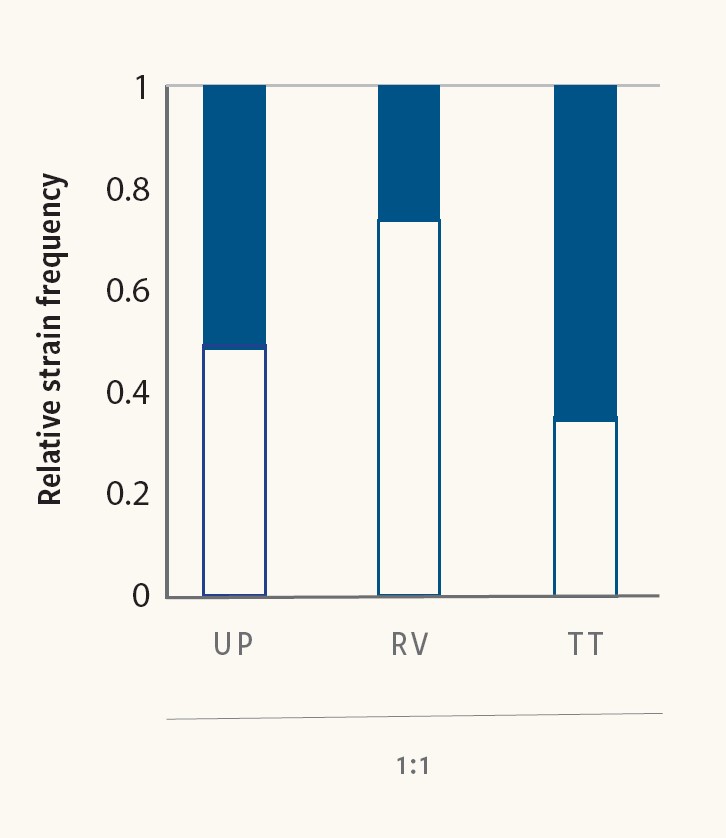
Figure 6. Mixed Salmonella cultures reveal competitive advantages between strains during pre‐enrichment and selective enrichment
Larsen et al. (2021) used CRISPR SeroSeq to demonstrate that the choice of culture media can influence the competitive dynamics between different strains. Figure 6 shows specific Salmonella Montevideo (white) and Salmonella Typhimurium (black) strains that were mixed at equal ratios and cultured in two different broths. In Rappaport-Vassiliadis soya peptone broth, the S. Montevideo strain outgrew the S. Typhimurium strain, whereas in Tetrathionate broth, the S. Typhimurium strain outgrew the S. Montevideo strain.
Antimicrobial susceptibility among Salmonella strains
Antimicrobial susceptibility is another important consideration in implementing a Salmonella-intervention process in live production.
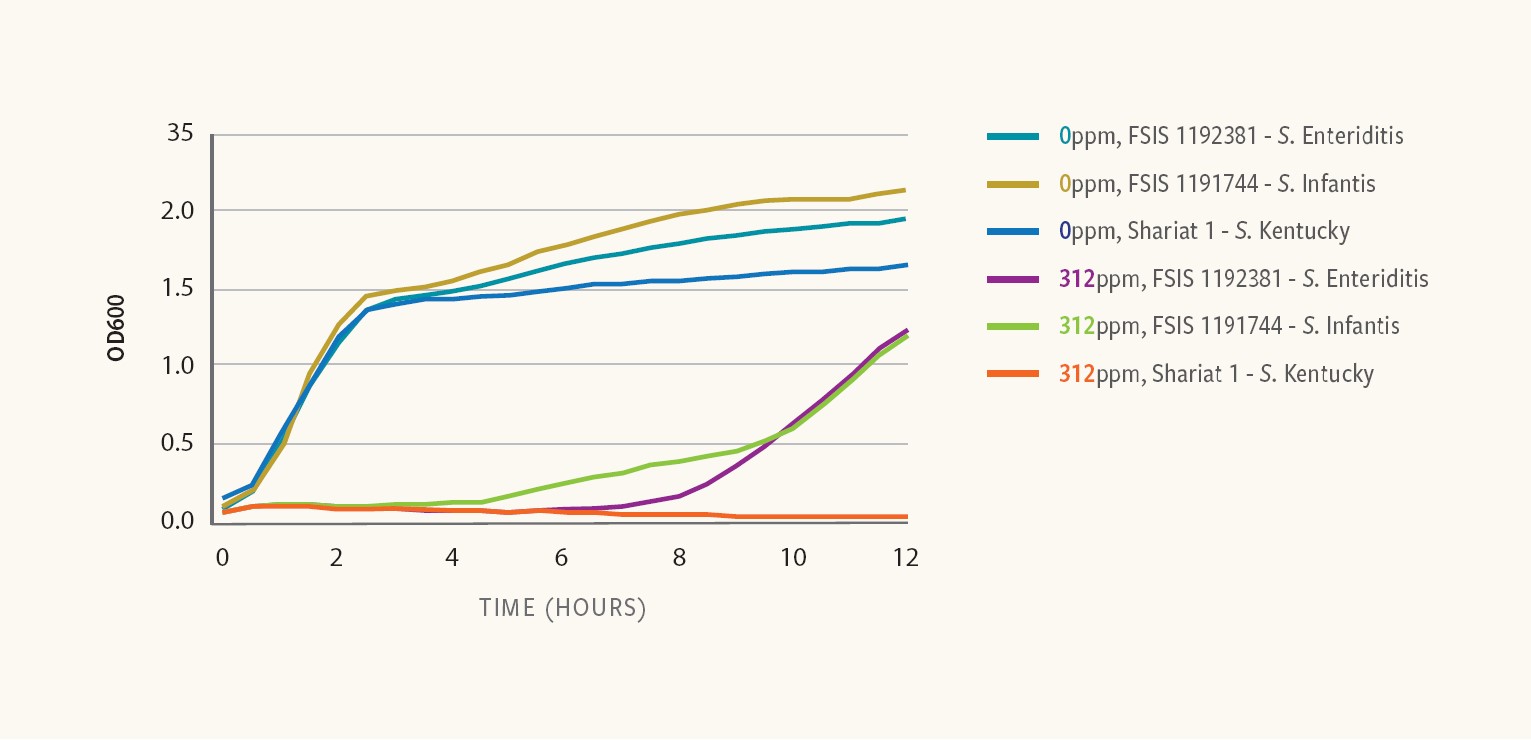
Figure 7. Antimicrobial susceptibility of S. Enteriditis, S. Infantis and S. Kentucky
Ancera’s research showed similar growth kinetics among three different Salmonella strains in rich media in the absence of any antimicrobial.
However, in the presence of 312 ppm peracetic acid (PAA) — an organic chemical used to control Salmonella in processing — the results revealed different responses among the strains, with some strains resuming growth shortly after exposure to PAA, while others remained unaffected even after extended incubation periods, Kiss says.
A strain of S. Infantis resumed growing within 4 hours of exposure, while a strain of S. Enteritidis resumed growing within 8 hours after exposure and a strain of S. Kentucky did not resume growth at all within 12 hours.
Salmonella monitoring at scale
Proper interpretation of Salmonella-surveillance results requires a deep understanding of growth kinetics, selective media and antimicrobial susceptibility. According to Kiss, there is inherent complexity within the variability in lag time, doubling time and response to selective media among different strains and serovars.
As food-safety professionals develop multi-layered food-defense systems that effectively detect, quantify, categorize and intervene against Salmonella, it is critical that they have granular serotype information and robust data systems to understand and explain the interconnected relationship between varying strains.
“Food producers looking for a more detailed understanding of their Salmonella-control operations will require interdisciplinary collaborations between microbiologists, epidemiologists and food-safety experts,” Kiss says.
“They will need to leverage advanced molecular biology, genomics and data analytics. They will need researchers who can further improve the sensitivity, specificity and efficiency of Salmonella-surveillance methods. Or they can work with Ancera, which does all of this and more.”
Editor’s note: Content on Modern Poultry’s Industry Insights pages is provided and/or commissioned by our sponsors, who assume full responsibility for its accuracy and compliance.